lXtractor.ext package
lXtractor.ext.base module
Base utilities for the ext module, e.g., base classes and common functions.
- class lXtractor.ext.base.ApiBase(url_getters, max_trials=1, num_threads=None, verbose=False)[source]
Bases:
objectBase class for simple APIs for webservices.
- __init__(url_getters, max_trials=1, num_threads=None, verbose=False)[source]
- Parameters:
url_getters (dict[str, UrlGetter]) – A dictionary holding functions constructing urls from provided args.
max_trials (int) – Max number of fetching attempts for a given query (PDB ID).
num_threads (int | None) – The number of threads to use for parallel requests. If
None,will send requests sequentially.verbose (bool) – Display progress bar.
- max_trials: int
Upper limit on the number of fetching attempts.
- num_threads: int | None
The number of threads passed to the
ThreadPoolExecutor.
- property url_args: list[tuple[str, list[str]]]
- Returns:
A list of services and argument names necessary to construct a valid url.
- url_getters: dict[str, UrlGetter]
A dictionary holding functions constructing urls from provided args.
- property url_names: list[str]
- Returns:
A list of supported services.
- verbose: bool
Display progress bar.
- class lXtractor.ext.base.StructureApiBase(url_getters, max_trials=1, num_threads=None, verbose=False)[source]
Bases:
ApiBaseA generic abstract API to fetch structures and associated info.
Child classes must implement
supported_str_formats()and have a url constructor named “structures” inurl_getters.- fetch_info(service_name, url_args, dir_, *, overwrite=False, callback=<function load_json_callback>)[source]
Fetch text information.
- Parameters:
service_name (str) – The name of the service to get a url_getter from
url_getters.dir – Dir to save files to. If
None, will keep downloaded files as strings.url_args (Iterable[_ArgT]) – Arguments to a url_getter.
overwrite (bool) – Overwrite existing files if dir_ is provided.
callback (Callable[[_ArgT, _RT], _T] | None) – Callback to apply after fetching the information file. By default, the content is assumed to be in
jsonformat. Thus, the default callback will parse the fetched content asdict. To disable this behavior, passcallback=None.
- Returns:
A tuple with fetched and remaining inputs. Fetched inputs are tuples, where the first element is the original arguments and the second argument is the dictionary with downloaded data. Remaining inputs are arguments that failed to fetch.
- Return type:
tuple[list[tuple[_ArgT, dict | Path]], list[_ArgT]]
- fetch_structures(ids, dir_, fmt='cif', *, overwrite=False, parse=False, callback=None)[source]
Fetch structure files.
PDB example:
See also
lXtractor.util.io.fetch_files().Hint
Callbacks will apply in parallel if
num_threadsis above 1.Note
If the provided callback fails, it is equivalent to the fetching failure and will be presented as such. Initializing in verbose mode will output the stacktrace.
Reading structures and parsing immediately requires using
callback. Such callback may be partially evaluatedlXtractor.core.structure.GenericStructure.read()encapsulating the correct format.- Parameters:
ids (Iterable[str]) – An iterable over structure IDs.
dir – Dir to save files to. If
None, will keep downloaded files as strings.fmt (str) – Structure format. See
supported_str_formats(). Adding .gz will fetch gzipped files.overwrite (bool) – Overwrite existing files if dir_ is provided.
parse (bool) – If
dir_ is None, useparse_callback(fmt=fmt)()to parse fetched structures right away. This will override any existing callback.callback (Callable[[tuple[str, str], _RT], _T] | None) – If dir_ is omitted, fetching will result in a
bytesor astr. Callback is a single-argument callable accepting the fetched content and returning anything.
- Returns:
A tuple with fetched results and the remaining IDs. The former is a list of tuples, where the first element is the original ID, and the second element is either the path to a downloaded file or downloaded data as string. The order may differ. The latter is a list of IDs that failed to fetch.
- Return type:
tuple[list[tuple[tuple[str, str], Path | _RT | _T]], list[tuple[str, str]]]
- abstract property supported_str_formats: list[str]
- Returns:
A list of formats supported by
fetch_structures().
- class lXtractor.ext.base.SupportsAnnotate(*args, **kwargs)[source]
Bases:
Protocol[CT]A class that serves as basis for annotators – callables accepting a Chain*-type object and returning a single or multiple objects derived from an initial Chain*, e.g., via
spawn_child <lXtractor.core.chain.Chain.spawn_child().- __init__(*args, **kwargs)
- lXtractor.ext.base.load_json_callback(_, res)[source]
- Parameters:
_ (Any) – Arguments to the
url_getter()(ignored).res (str) – Fetched string content.
- Returns:
Parsed json as
dict.- Return type:
dict
- lXtractor.ext.base.parse_structure_callback(inp, res)[source]
Parse the fetched structure.
- Parameters:
inp (tuple[str, str]) – A pair of (id, fmt).
res (str | bytes) – The fetching result. By default, if
fmt in ["cif", "pdb"], the result isstr, whilefmt="mmtf"will producebytes.
- Returns:
Parse generic structure.
- Return type:
lXtractor.ext.hmm module
Wrappers around PyHMMer for convenient annotation of domains and families.
- class lXtractor.ext.hmm.Pfam(resource_path=PosixPath('/home/docs/checkouts/readthedocs.org/user_builds/lxtractor/checkouts/latest/lXtractor/resources/Pfam'), resource_name='Pfam')[source]
Bases:
AbstractResourceA minimalistic Pfam interface.
Parsed Pfam data is represented as a Pandas DataFrame accessible via
df()with columns: “ID”, “Accession”, “Description”, “Category”, and “HMM”. Each row corresponds to a single model from Pfam-A collection and associated metadata taken from the Pfam-A.dat file. HMM models are wrapped into aPyHMMerinstance.For quick access to a single HMM model parsed into
PyHMMer, usePfam()[hmm_id].- __init__(resource_path=PosixPath('/home/docs/checkouts/readthedocs.org/user_builds/lxtractor/checkouts/latest/lXtractor/resources/Pfam'), resource_name='Pfam')[source]
- Parameters:
resource_path (Path) – Path to parsed resource data.
resource_name (str) – Resource’s name.
- clean(raw=True, parsed=False)[source]
Remove Pfam data. If raw and parsed are both
False, removes thepathwith all stored data.- Parameters:
raw (bool) – Remove raw fetched files.
parsed (bool) – Remove parsed files.
- Returns:
Nothing.
- Return type:
None
- dump(path=None)[source]
Store parsed data to the filesystem.
This function will store the HMM metadata to attr:path / “parsed” / “dat.csv” and separate gzip-compressed HMM models into
path/ “parsed” / “hmm”.- Parameters:
path (Path | None) – Use this path instead of the
pathas a base dir.- Returns:
The path
path/ “parsed”.- Return type:
Path
- fetch(url_hmm='https://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.gz', url_dat='https://ftp.ebi.ac.uk/pub/databases/Pfam/current_release/Pfam-A.hmm.dat.gz')[source]
Fetch Pfam-A data from InterPro.
- Parameters:
url_hmm (str) – URL to “Pfam-A.hmm.gz”.
url_dat (str) – URL to “Pfam-A.hmm.dat.gz”
- Returns:
A pair of filepaths for fetched HMM and dat files.
- Return type:
tuple[Path, Path]
- load_hmm(df=None, path=None)[source]
Load HMM models according to accessions in passed df and create a column “PyHMMer” with loaded models.
- Parameters:
df (DataFrame | None) – A
DataFramehaving all the :meth:`dat_columns.path (Path | None) – A custom path to the parsed data with an “hmm” subdir.
- Returns:
A copy of the original
DataFramewith loaded models.- Return type:
DataFrame
- parse(dump=True, rm_raw=True)[source]
Parse fetched raw data into a single pandas
DataFrame.- Parameters:
dump (bool) – Dump parsed files to
path/ “raw” dir.rm_raw (bool) – Clean up the raw data once parsing is done.
- Returns:
A parsed Pfam
DataFrame. See the class’s docs for a list of columns.- Return type:
DataFrame
- read(path=None, accessions=None, categories=None, hmm=True)[source]
Read parsed Pfam data.
First it reads the “dat” file and filters to relevant accessions and/or categories. Then, if hmm is
True, it loads each model and wraps into anPyHMMerinstance. Otherwise, it loads the HMM metadata. One can explore and filter these data, then load the desired HMM models viaload_hmm().- Parameters:
path (Path | None) – A path to the dir with layout similar to what
dump()creates.accessions (Container[str] | None) – A list of Pfam accessions following the “.”, e.g.,
["PF00069", ].categories (Container[str] | None) – A list of Pfam categories to filter the accessions to.
hmm (bool) – Load HMM models.
- Returns:
A parsed Pfam
DataFrame.- Return type:
DataFrame
- property dat_columns: tuple[str, ...]
- class lXtractor.ext.hmm.PyHMMer(hmm, **kwargs)[source]
Bases:
objectA basis pyhmmer interface aimed at domain extraction. It works with a single hmm model and pipeline instance.
The original documentation <https://pyhmmer.readthedocs.io/en/stable/>.
- __init__(hmm, **kwargs)[source]
- Parameters:
hmm (HMM | HMMFile | Path | str) – An
HMMFilehandle or path as string or Path object to a file containing a single HMM model. In case of multiple models, only the first one will be takenkwargs – Passed to
Pipeline. The alphabet argument is derived from the supplied hmm.
- align(seqs)[source]
Align sequences to a profile.
- Parameters:
seqs (Iterable[Chain | ChainStructure | ChainSequence | str | tuple[str, str] | DigitalSequence]) – Sequences to align.
- Returns:
TextMSAwith aligned sequences.- Return type:
TextMSA
- annotate(objs, new_map_name=None, min_score=None, min_size=None, min_cov_hmm=None, min_cov_seq=None, domain_filter=None, **kwargs)[source]
Annotate provided objects by hits resulting from the HMM search.
An annotation is the creation of a child object via
spawn_child()method (e.g.,lXtractor.core.chain.ChainSequence.spawn_child()).- Parameters:
objs (Iterable[Chain | ChainStructure | ChainSequence] | Chain | ChainStructure | ChainSequence) – A single one or an iterable over Chain*-type objects.
new_map_name (str | None) – A name for a child
ChainSequence <lXtractor.core.chain.ChainSequenceto hold the mapping to the hmm numbering.min_score (float | None) – Min hit score.
min_size (int | None) – Min hit size.
min_cov_hmm (float | None) – Min HMM model coverage – a fraction of mapped / total nodes.
min_cov_seq (float | None) – Min coverage of a sequence by the HMM model nodes – a fraction of mapped nodes to the sequence’s length.
domain_filter (Callable[[Domain], bool] | None) – A callable to filter domain hits.
kwargs – Passed to the spawn_child method. Hint: if you don’t want to keep spawned children, pass
keep=Falsehere.
- Returns:
A generator over spawned children yielded sequentially for each input object and valid domain hit.
- Return type:
Generator[CT, None, None]
- convert_seq(obj)[source]
- Parameters:
obj (Any) – A Chain*-type object or string or a tuple of (name, _seq). A sequence of this object must be compatible with the alphabet of the HMM model.
- Returns:
A digitized sequence compatible with PyHMMer.
- Return type:
DigitalSequence
- classmethod from_hmm_collection(hmm, **kwargs)[source]
Split HMM collection and initialize a
PyHMMerinstance from each HMM model.- Parameters:
hmm (_HmmInpT) – A path to HMM file, opened HMMFile handle, or parsed HMM.
kwargs – Passed to the class constructor.
- Returns:
A generator over
PyHMMerinstances created from the provided HMM models.- Return type:
abc.Generator[t.Self]
- classmethod from_msa(msa, name, alphabet, **kwargs)[source]
Create a
PyHMMerinstance from a multiple sequence alignment.- Parameters:
msa (abc.Iterable[tuple[str, str] | str | _ChainT] | lXAlignment) – An iterable over sequences.
name (str | bytes) – The HMM model’s name.
alphabet (Alphabet | str) – An alphabet to use to build the HMM model. See
digitize_seq()for available options.kwargs – Passed to
DigitalMSAofPyHMMerthat serves as the basis for creating an HMM model.
- Returns:
A new
PyHMMerinstance initialized with the HMM model built here.- Return type:
t.Self
- init_pipeline(**kwargs)[source]
- Parameters:
kwargs – Passed to
Pipelineduring initialization.- Returns:
Initialized pipeline, also saved to
pipeline.- Return type:
Pipeline
- search(seqs)[source]
Run the
pipelineto search forhmm.- Parameters:
seqs (Iterable[Chain | ChainStructure | ChainSequence | str | tuple[str, str] | DigitalSequence]) – Iterable over digital sequences or objects accepted by
convert_seq().- Returns:
Top hits resulting from the search.
- Return type:
TopHits
- hits_: TopHits | None
Hits resulting from the most recent HMM search
- hmm
HMM instance
- pipeline: Pipeline
Pipeline to use for HMM searches
- lXtractor.ext.hmm.digitize_seq(obj, alphabet='amino')[source]
- Parameters:
obj (Any) – A Chain*-type object or string or a tuple of (name, _seq). A sequence of this object must be compatible with the alphabet of the HMM model.
alphabet (Alphabet | str) – An alphabet type the sequence corresponds to. Can be an initialized PyHMMer alphabet or a string “amino”, “dna”, or “rna”.
- Returns:
A digitized sequence compatible with PyHMMer.
- Return type:
DigitalSequence
lXtractor.ext.pdb_ module
Utilities to interact with the RCSB PDB database.
- class lXtractor.ext.pdb_.PDB(max_trials=1, num_threads=None, verbose=False)[source]
Bases:
StructureApiBaseBasic RCSB PDB interface to fetch structures and information.
Example of fetching structures:
>>> pdb = PDB() >>> fetched, failed = pdb.fetch_structures(['2src', '2oiq'], dir_=None) >>> len(fetched) == 2 and len(failed) == 0 True >>> (args1, res1), (args2, res2) = fetched >>> assert {args1, args2} == {('2src', 'cif'), ('2oiq', 'cif')} >>> isinstance(res1, str) and isinstance(res2, str) True
Example of fetching information:
>>> pdb = PDB() >>> fetched, failed = pdb.fetch_info( ... 'entry', [('2SRC', ), ('2OIQ', )], dir_=None) >>> len(failed) == 0 and len(fetched) == 2 True >>> (args1, res1), (args2, res2) = fetched >>> assert {args1, args2} == {('2SRC', ), ('2OIQ', )} >>> assert isinstance(res1, dict) and isinstance(res2, dict)
Hint
Check
list_services()to list available info services.- __init__(max_trials=1, num_threads=None, verbose=False)[source]
- Parameters:
url_getters – A dictionary holding functions constructing urls from provided args.
max_trials (int) – Max number of fetching attempts for a given query (PDB ID).
num_threads (int | None) – The number of threads to use for parallel requests. If
None,will send requests sequentially.verbose (bool) – Display progress bar.
- static fetch_obsolete()[source]
- Returns:
A dict where keys are obsolete PDB IDs and values are replacement PDB IDs or an empty string if no replacement was made.
- Return type:
dict[str, str]
- property supported_str_formats: list[str]
- Returns:
A list of formats supported by
fetch_structures().
- lXtractor.ext.pdb_.filter_by_method(pdb_ids, pdb=<lXtractor.ext.pdb_.PDB object>, method='X-ray', dir_=None)[source]
See also
PDB.fetch_infoNote
Keys for the info dict are ‘rcsb_entry_info’ -> ‘experimental_method’
- Parameters:
pdb_ids (Iterable[str]) – An iterable over PDB IDs.
pdb (PDB) – Fetcher instance. If not provided, will init with default params.
method (str) – Method to match. Must correspond exactly.
dir – Dir to save info “entry” json dumps.
- Returns:
A list of PDB IDs obtained by desired experimental procedure.
- Return type:
list[str]
lXtractor.ext.sifts module
Contains utils allowing to benefit from SIFTS database UniProt-PDBF; mapping.
Namely, the SIFTS class is build around the file uniprot_segments_observed.csv.gz. The latter contains segment-wise mapping between UniProt sequences and continuous corresponding regions in PDB structures, and allows us to:
#. Cross-reference PDB and UniProt databases (e.g., which structures are available for a UniProt “PXXXXXX” accession?) #. Map between sequence numbering schemes.
- class lXtractor.ext.sifts.Mapping(id_from, id_to, *args, **kwargs)[source]
Bases:
UserDictA
dictsubclass with explicit IDs of keys/values sources.
- class lXtractor.ext.sifts.SIFTS(resource_path=None, resource_name='SIFTS', load_segments=False, load_id_mapping=False)[source]
Bases:
AbstractResourceA resource to segment-wise and ID mappings between UniProt and PDB.
For a first-time usage, you’ll need to call
fetch()to download and store the “uniprot_segments_observed” dataset.>>> sifts = SIFTS() >>> path = sifts.fetch() >>> path.name 'uniprot_segments_observed.csv.gz'
Next,
parse()will process the downloaded file to create and store the table with segments and ID mappings.(We pass
overwrite=Truefor the doctest to work. It’s not needed for the first setup).>>> df, mapping = sifts.parse(store_to_resources=True, overwrite=True) >>> isinstance(df, pd.DataFrame) and isinstance(mapping, dict) True >>> list(df.columns)[:4] ['PDB_Chain', 'PDB', 'Chain', 'UniProt_ID'] >>> list(df.columns)[4:] ['PDB_start', 'PDB_end', 'UniProt_start', 'UniProt_end']
Now that we parsed SIFTS segments data, we can use it to map IDs and numberings between UniProt and PDB. Let’s reinitalize SIFTS to verify it loads locally stored resources
>>> sifts = SIFTS(load_segments=True, load_id_mapping=True) >>> assert isinstance(sifts.df, pd.DataFrame) >>> assert isinstance(sifts.id_mapping, dict)
SIFTS has three types of mappings stored:
Between UniProt and PDB Chains
>>> sifts['P12931'][:4] ['1A07:A', '1A07:B', '1A08:A', '1A08:B']
Between PDB Chains and UniProt IDs
>>> sifts['1A07:A'] ['P12931']
Between PDB IDs and PDB Chains
>>> sifts['1A07'] ['A', 'B']
The same types of keys are supported to obtain mappings between the numbering schemes. You’ll get a generator yielding mappings from UniProt numbering to the PDB numbering.
In these two cases, we’ll get the mappings for each chain.
>>> mappings = list(sifts.map_numbering('P12931')) >>> assert len(mappings) == len(sifts['P12931']) >>> mappings = list(sifts.map_numbering('1A07')) >>> assert len(mappings) == len(sifts['1A07']) == 2
If we specify the chain, we get a single mapping.
>>> m = next(sifts.map_numbering('1A07:A')) >>> list(m.items())[:2] [(145, 145), (146, 146)]
- __init__(resource_path=None, resource_name='SIFTS', load_segments=False, load_id_mapping=False)[source]
- Parameters:
resource_path (Path | None) – a path to a file “uniprot_segments_observed”. If not provided, will try finding this file in the
resourcesmodule. If the latter fails will attempt fetching the mapping from the FTP server and storing it in theresourcesfor later use.resource_name (str) – the name of the resource.
load_segments (bool) – load pre-parsed segment-level mapping
load_id_mapping (bool) – load pre-parsed id mapping
- dump(path, **kwargs)[source]
- Parameters:
path (Path) – a valid writable path.
kwargs – passed to
DataFrame.to_csv()method.
- Returns:
- fetch(url='ftp://ftp.ebi.ac.uk/pub/databases/msd/sifts/flatfiles/csv/uniprot_segments_observed.csv.gz', overwrite=False)[source]
Download the resource.
- static load()[source]
- Returns:
Loaded segments df and name mapping or
Noneif they don’t exist.- Return type:
tuple[DataFrame | None, dict[str, list[str]] | None]
- map_id(x)[source]
- Parameters:
x (str) – Identifier to map from.
- Returns:
A list of IDs that x maps to.
- Return type:
list[str] | None
- map_numbering(obj_id)[source]
Retrieve mappings associated with the
obj_id. Mapping example:1 -> 2 2 -> 3 3 -> None 4 -> 4
Above, a UniProt sequence maps to two segments of a PDB sequence (2-3 and 4). PDB sequence is always considered a subset of a corresponding UniProt sequence. Thus, any “holes” between continuous PDB segments are filled with
None.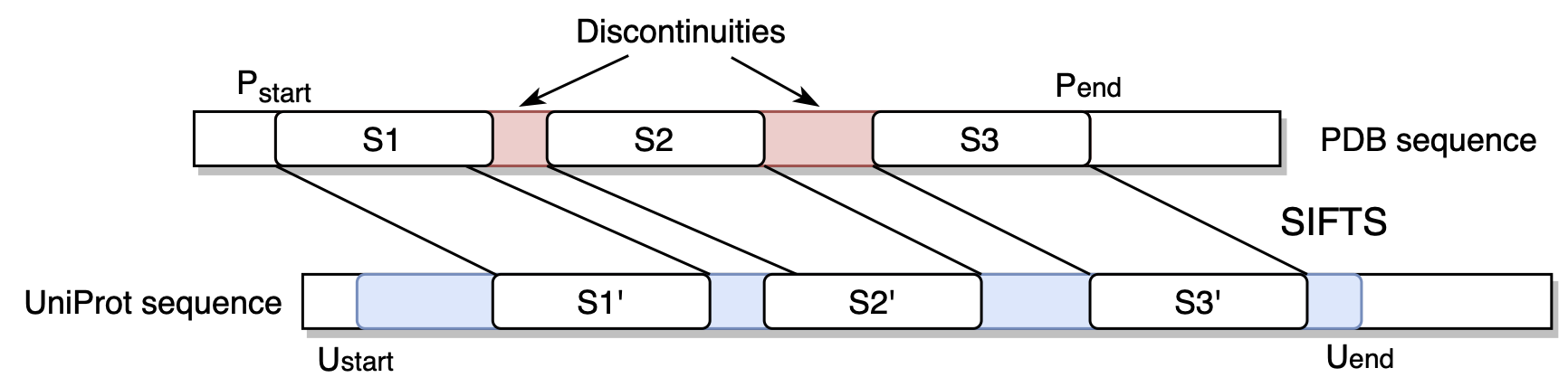
Mapping from PDB segments to UniProt segments accounting for discontinuities.
- Parameters:
obj_id (str) –
a string value in three possible formats:
”PDB ID:Chain ID”
”PDB ID”
”UniProt ID”
- Returns:
an iterator over the
Mappingobjects. These are “unidirectional”, i.e., theMappingis always from the UniProt numbering to the PDB numbering regardless of theobj_idnature.- Return type:
Generator[Mapping]
- parse(overwrite=False, store_to_resources=True, rm_raw=True)[source]
Prepare the resource to be used for mapping:
remove records with empty chains.
- select and rename key columns based on the
SIFTS_RENAMES constant.
- select and rename key columns based on the
create a PDB_Chain column to speed up the search.
- Parameters:
overwrite (bool) – Overwrite both
dfand existing id mapping and parsed segments.store_to_resources (bool) – Store parsed DataFrame and id mapping in resources for further simplified access.
rm_raw (bool) – After parsing is finished, remove raw SIFTS download. (!) If `store_to_resources` is ``False``, using SIFTS next time will require downloading “uniprot_segments_observed”.
- Returns:
prepared
DataFrameof Segment-wise mapping between UniProt and PDB sequences. Mapping between IDs will be stored inid_mapping.- Return type:
tuple[DataFrame, dict[str, list[str]]]
- prepare_mapping(up_ids: Iterable[str], pdb_ids: Iterable[str] | None = None, pdb_method: str | None = 'X-ray', pdb_base: Path | None = None, pdb_fmt: str = 'cif', pdb_method_filter_kwargs: Mapping[str, Any] | None = None) Mapping[str, list[tuple[str | Path, list[str]]]][source]
- prepare_mapping(up_ids: Mapping[str, _Mkey], pdb_ids: Iterable[str] | None = None, pdb_method: str | None = 'X-ray', pdb_base: Path | None = None, pdb_fmt: str = 'cif', pdb_method_filter_kwargs: Mapping[str, Any] | None = None) Mapping[_Mkey, list[tuple[str | Path, list[str]]]]
- prepare_mapping(up_ids: Iterable[str] | Mapping[str, _Mkey], pdb_ids: Iterable[str] | None = None, pdb_method: str | None = 'X-ray', pdb_base: None = None, pdb_fmt: str = 'cif', pdb_method_filter_kwargs: Mapping[str, Any] | None = None) Mapping[str | _Mkey, list[tuple[str, list[str]]]]
- prepare_mapping(up_ids: Iterable[str] | Mapping[str, _Mkey], pdb_ids: Iterable[str] | None = None, pdb_method: str | None = 'X-ray', pdb_base: Path = None, pdb_fmt: str = 'cif', pdb_method_filter_kwargs: Mapping[str, Any] | None = None) Mapping[str | _Mkey, list[tuple[Path, list[str]]]]
Prepare mapping to use with
lXtractor.core.chain.initializer.ChainInitializer.from_mapping().Uses SIFTS’ UniProt-PDB mappings to derive mapping of the form:
UniProtID => [(PDB code, [PDB chains]), ...]
- Parameters:
up_ids – UniProt IDs to map with
SIFTSor a mapping of UniProt IDs to objects allowed as keys infrom_mapping().pdb_ids – PDB IDs to restrict the mapping to. Can be regular IDs or with chain specifier (eg “1ABC:A”).
pdb_method – Filter PDB IDs by experimental method.
pdb_base –
A path to a PDB files’ dir. If provided, the mapping takes the form:
UniProtID => [(PDB path, [PDB chains]), ...]
pdb_fmt – PDB file format for files in pdb_base.
pdb_method_filter_kwargs – A keyword arguments passed to
lXtractor.ext.pdb_.filter_by_method()used to filter PDB IDs.
- Returns:
A mapping that is almost ready to be used with
lXtractor.core.chain.initializer.ChainInitializer. The only preparation step left is to replace the keys with compatible type.
- read(overwrite=True)[source]
The method reads the initial file “uniprot_segments_observed” into memory.
To load parsed files, use
load().- Parameters:
overwrite (bool) – overwrite existing
dfattribute.- Returns:
pandas
DataFrameobject.- Return type:
DataFrame
- property pdb_chains: set[str]
- Returns:
A set of encompassed PDB Chains (in {PDB_ID}:{PDB_Chain} format).
- property pdb_ids: set[str]
- Returns:
A set of encompassed PDB IDs.
- property uniprot_ids: set[str]
- Returns:
A set of encompassed UniProt IDs.
lXtractor.ext.uniprot module
- class lXtractor.ext.uniprot.UniProt(chunk_size=100, max_trials=1, num_threads=1, verbose=False)[source]
Bases:
ApiBaseAn interface to UniProt fetching.
UniProt.url_gettersdefines functions that construct a URL from provided arguments to fetch specific data. For instance, calling a URL getter for sequences in fasta format using a list of sequences will construct a valid URL for fetching the data.>>> uni = UniProt() >>> uni.url_getters['sequences'](['P00523', 'P12931']) 'https://rest.uniprot.org/uniprotkb/stream?format=fasta&query=accession%3AP00523+OR+accession%3AP12931'
These URLs are constructed dynamically within this class’s methods, used to query UniProt, fetch and parse the data.
- __init__(chunk_size=100, max_trials=1, num_threads=1, verbose=False)[source]
- Parameters:
chunk_size (int) – A number of IDs to join within a single URL and query simultaneously. Note that having invalid URL in a chunk invalidates all its IDs: they won’t be fetched. For optimal performance, please filter your accessions carefully.
max_trials (int) – A maximum number of trials for fetching a single chunk. Makes sense to raise above
1when the connection is unstable.num_threads (int) – The number of threads to use for fetching chunks in parallel.
verbose (bool) – Display progress bar via stdout.
- fetch_info(accessions, fields=None, as_df=True)[source]
Fetch information in tsv format from UniProt.
- Parameters:
accessions (Iterable[str]) – A list of accessions to fetch the info for.
fields (str | None) – A comma-separated list of fields to fetch. If
None, default fields UniProt provides will be used.as_df (bool) – Convert fetched tables into pandas dataframes and join them. Otherwise, return raw text corresponding to each chunk of accessions.
- Returns:
A list of texts per chunk or a single data frame.
- Return type:
DataFrame | list[str]
- fetch_sequences(accessions, dir_, overwrite, callback: None) Iterator[tuple[str, str]][source]
- fetch_sequences(accessions, dir_, overwrite, callback: Callable[[tuple[str, str]], T]) Iterator[T]
Fetch sequences in “fasta” format from UniProt.
- Parameters:
accessions – A list of valid accessions to fetch.
dir – A directory where individual sequence will be stored. If exists, will filter accessions before fetching unless overwrite is
True.overwrite – Overwrite existing sequences if they exist in dir_.
callback – A function accepting a single sequence and returning anything else. Can be useful to convert sequences into, eg, :class:~lXtractor.chain.sequence.ChainSequence` (for this, pass :meth:~lXtractor.chain.sequence.ChainSequence.from_tuple` here).
- Returns:
An iterator over fetched sequences (or whatever
callbackreturns).
- lXtractor.ext.uniprot.fetch_uniprot(acc, fmt='fasta', chunk_size=100, fields=None, **kwargs)[source]
An interface to the UniProt’s search.
Base URL: https://rest.uniprot.org/uniprotkb/stream
Available DB identifiers: See bioservices <https://bioservices.readthedocs.io/en/main/_modules/bioservices/uniprot.html>
- Parameters:
acc (Iterable[str]) – an iterable over UniProt accessions.
fmt (str) – download format (e.g., “fasta”, “gff”, “tab”, …).
chunk_size (int) – how many accessions to download in a chunk.
fields (str | None) – if the
fmtis “tsv”, must be provided to specify which data columns to fetch.kwargs – passed to
fetch_chunks().
- Returns:
the ‘utf-8’ encoded results as a single chunk of text.
- Return type:
str